Quite shockingly(not!), Opisthokont got the last one. I agree with his statement that that was like shooting fish in a barrel, but easier since fish are actually difficult to shoot at from air due to refraction, etc. The organism behind the shell in the mystery micrograph was...
Euglypha!

(Wikipedia; Euglyphid test)
Euglypha morphologically belongs to the polyphyletic 'testate amoebae', but is phylogenetically quite distant from your garden variety test-building amoebozoans, like Arcella and Difflugia. Euglyphids are cercozoan rhizarians. Since those words likely mean nothing to the vast majority of the human population, here's a 'map' of Rhizaria in my Coelodiceras (Phaeodaria) post, and an overall tree of eukaryotes can be found here. They can often be found in moss samples, but are also present in soil and freshwater environments. Their test scales are made of silica, and preserve quite decently as fossils. Speaking of which, I apparently may have a slight fetish for unicellular microfossils:
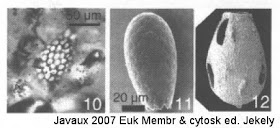
(Javaux 2007 in Eukaryotic Membranes and Cytoskeleton: Origins and Evolution ed. Jékely; 10 - fossil; 11 - modern Euglyphid; 12 - VSM - 'vase-shaped microfossil' (micropaleontological equivalent of mycologists' LBM - 'little brown mushroom' ?) with holes possibly caused by predation; fossils ~750My old)
Fast forward 750My back to the present, the modern euglyphids are about as diverse as they are understudied:

(Lara et al. 2007 Protist; tree of Euglyphids)
Images of Euglyphid diversity, shamelessly stolen from the same paper:

(Lara et al. 2007 Protist; Euglyphid SEMs, scalebar 50um except for E,F,H, where it's 10um. A - Assulina; B- Placocista; C,D - Euglypha ciliata & compressa; E - Corythion; F - Trinema; H,G - Euglypha penardi)
Unfortunately, finding nice plates full of euglyphids is rather difficult, since until quite recently, they were lumped together with testate Amoebozoans. Also, since euglyphids fossilise, they seem to be mostly studied by paleontologists, who seem to have an 'interesting' relationship with systematics of the living. Where 'interesting' entails being at least a couple decades out of date. Well, they are millions of years in the past...
Paulinella can be argued to be particularly interesting - it is a case of an independent event of primary plastid endosymbiosis. Why this is interesting can be seen in this really nice overview:

(Keeling 2oo4 Am J Bot (free access); overview of plastid endosymbiosis - the 'Pacmen' are pretty awesome! Interestingly, if the Chromalveolate Hypothesis is correct, this would mean that Paulinella already had a plastid in its ancient past. However, it would've been a red algal plastid of a different cyanobacterial origin, not a Synechococcus-derived cyanelle)
Cyanelles are photosynthetic endosymbionts/organelles - they differ from plastids by retaining some prokaryotic features like the peptidoglycan wall. Among the conventional plastid-bearing algae, glaucophytes carry cyanelles from the primary cyanobacterial endosymbiotic event which eventually led to plant chloroplasts and most algal plastids. In sequence comparisons, Paulinella cyanelles branch with the cyanobacterium Synechococcus, and retain much of the gene order, suggesting a fairly recent endosymbiosis with Synechococcus (Yoon et al. 2006 Curr Biol). The authors predict a transfer of plastid division genes to the host nucleus; however, while the plastid genome has been sequenced (Nowack et al. 2008 Curr Biol), the nuclear genome is yet to be investigated, to my knowledge.

(Lauderborn 1895 (L) Melkonian (R) in Keeling & Archibald 2008 Curr Biol; Paulinella with cyanelles)
So are the Paulinella cyanelles to be considered as endosymbionts or organelles? As with the Perkinsela case discussed a while ago, who cares? Keeling & Archibald (2008) and Bodył et al. (2007) argue that the distinction between organelles and endosymbionts is too vague and fuzzy to obsess over, while Theissen & Martin (2006) seem to have nothing better to do. Well, to be fair, they argue that gene transfer to the host nucleus is the necessary rite of passage to become an organelle. But that is a rather arbitrary cut off, since you can also say that a complete disappearance of a certain class of genes, or the endosymbiont genome altogether, are necessary to be called an organelle. Alternatively, you could also argue that as soon as an endosymbiont spends its entire life cycle within the host cell, it's sufficient to be called an organelle. With lateral gene transfer turning out to be more ubiquitous than it first seemed, perhaps gene transfer isn't that significant of an event after all. Meanwhile, how about we just accept that nature doesn't particularly care for the artefacts of our reasoning -- ie. the obsessive-compulsive yearning to categorise the world around us -- and just enjoy the biology?
Nowack et al. (2008) avoid the whole Paulinella organelle vs. endosymbiont debate by settling for 'chromatophore'.
But just to annoy fellow cell biologists, I sometimes insist on referring to plastids and mitochondria as 'endosymbionts'. Speaking of which, from Theissen & Martin (2006):
"Calling the Paulinella endosymbiont a plastid or an organelle might make a story more exciting, but at the cost of scientific accuracy. Some proteobacterial endosymbionts of aphids have genomes smaller than those of some plastids [16]. Would anyone call those endosymbionts ‘mitochondria’? Hardly."First of all, why 'mitochondria'?! Those are specifically defined as endosymbionts/organelles of that one alpha-proteobacterial endosymbiosis event ancestral to all known eukaryotes. Second of all, I would totally call them organelles. I see no problem with it. I guess I'm just special then, according to those guys. Actually, I'll go around intentionally calling them 'organelles' just to piss them off.
So mitochondria and cyanobacterial plastids = endosymbionts; Paulinella cyanelles and Wolbachia bacterial endosymbionts = organelles. I think that pisses off just about everyone. My job here is done. =D
I hope this post has helped cast some familiarity upon yet another obscure group of Rhizarians. There's probably volumes more to say about them, but [insert rant about biomed crushing real biology here], so they're as understudied as the rest of Rhizaria. And much of the rest of Eukarya. Who knows how much biology weirdness lurks behind some of those obscure taxon names. Perhaps that 'Obscurius obscura' may hold the perfect properties to help us sort out some biological connundrum or other. Like what ciliate genomic madness has done for the discovery of telomeres (Blackburn & Gall 1978) and telomerase (Greider & Blackburn 1985). Come on guys, let's do something!
What if each lab was to take on a (culturable) obscure organism as a small side project? Even the big biomed labs... if each scientist played with a random organism long enough, perhaps we could unearth a freaking Pandora's box-worth of discoveries and new research directions? And the mixing of disciplines would do wonders to our overall understanding of biology! This may even be *gasp* slightly more efficient than 10 labs staring at one protein and barricading their labs from rampant scoopage... but I'll stop here with my heretic thoughts.
I have several posts in the works at the moment, among them one on origins of eukaryotes and a series on Heterolobosea, which Christopher Taylor challenged be to blog about. But first, a couple midterms, a talk I'm giving on Tuesday which I have yet to start working on, and a whole wad of research- and seminar course-related stuff. Learning seems to happen predominantly outside of class, which I find rather debilitating to grades. So multitasking must happen, and generally the class-related stuff gets cut. Which is bad. There's a dilemma between developing your mind/knowledge and getting decent marks - the former is essential for any reasonable progress in research/other professions, the latter is necessary to actually get anywhere after graduating. And they conflict with each other. Yay. [/rant]
And I seriously intended this post to be like "Hi, this is Euglypha. It is pretty. Gotta go. Bye." Cannot resist the allure of journal surfing at odd hours of the night...
References
BODYL, A., MACKIEWICZ, P., & STILLER, J. (2007). The intracellular cyanobacteria of Paulinella chromatophora: endosymbionts or organelles? Trends in Microbiology, 15 (7), 295-296 DOI: 10.1016/j.tim.2007.05.002
Javaux EJ (2007). The early eukaryotic fossil record. Advances in experimental medicine and biology, 607, 1-19 PMID: 17977455
Keeling, P. (2004). Diversity and evolutionary history of plastids and their hosts American Journal of Botany, 91 (10), 1481-1493 DOI: 10.3732/ajb.91.10.1481
KEELING, P., & ARCHIBALD, J. (2008). Organelle Evolution: What's in a Name? Current Biology, 18 (8) DOI: 10.1016/j.cub.2008.02.065
LARA, E., HEGER, T., MITCHELL, E., MEISTERFELD, R., & EKELUND, F. (2007). SSU rRNA Reveals a Sequential Increase in Shell Complexity Among the Euglyphid Testate Amoebae (Rhizaria: Euglyphida) Protist, 158 (2), 229-237 DOI: 10.1016/j.protis.2006.11.006
NOWACK, E., MELKONIAN, M., & GLOCKNER, G. (2008). Chromatophore Genome Sequence of Paulinella Sheds Light on Acquisition of Photosynthesis by Eukaryotes Current Biology, 18 (6), 410-418 DOI: 10.1016/j.cub.2008.02.051
THEISSEN, U., & MARTIN, W. (2006). The difference between organelles and endosymbionts Current Biology, 16 (24) DOI: 10.1016/j.cub.2006.11.020
Yoon, H., Reyes-Prieto, A., Melkonian, M., & Bhattacharya, D. (2006). Minimal plastid genome evolution in the Paulinella endosymbiont Current Biology, 16 (17) DOI: 10.1016/j.cub.2006.08.018

The fossils are actually from Porter et al., 2003 in Journal of Paleontology v77. Loving your blog!
ReplyDelete